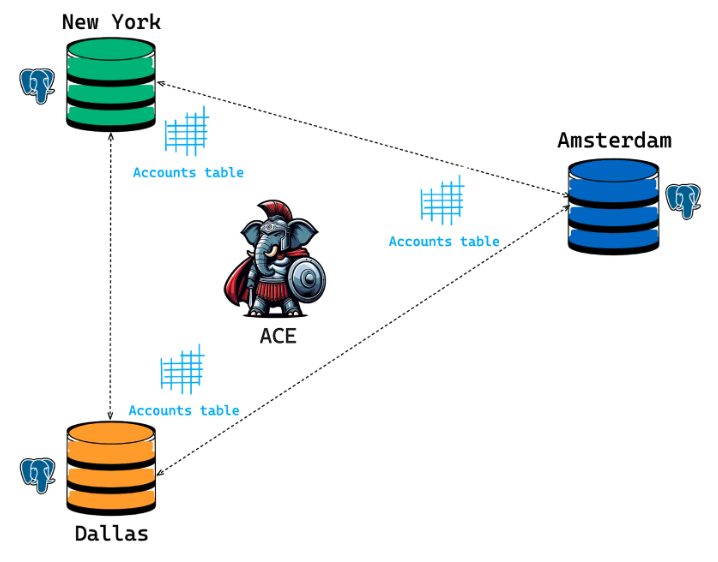
ACE (the Anti-Chaos Engine)
In an eventually consistent system (like a pgEdge cluster), your nodes have the potential to go out of sync (diverge) due to replication lag, network partitioning, or node failure. Database differences can also arise out of planned or unplanned downtime or unexpected cloud provider failures. The Anti Chaos Engine (ACE) allows for reconciliation of such differences across databases by performing efficient table comparisons and reporting found differences in output formats that are both machine and user friendly. You can also use ACE to repair the discrepancies by specifying the node that is considered your source-of-truth and instructing ACE to update the differences between the nodes.
Common Use Cases
Planned maintenance: If you need to take a database node down for administrative purposes, its state may fall behind the state of other nodes. When you bring the node online again, you can use ACE to compare node states and bring the node up to speed.
Node failures: Network failure can cause unplanned node downtime; use ACE to repair the node’s state and return it to the cluster.
Network Partition: In a typical deployment of pgEdge, a database cluster is often spread across multiple regions and data centers. Due to the volatility that comes with a large-scale deployment, network partitions are common. You can use ACE to return a cluster to a consistent state if the state is disrupted due to partition-related irregularities.
ACE Deployment Considerations
ACE is very efficient when it comes to comparing tables. It first looks at the table sizes and then based on the specified runtime options, splits the task into multiple processes and executes them in parallel. How fast it executes a table-diff command depends on the:
- configuration of the machine ACE is being run on – how many cores, how much memory, etc.
- size of your table.
- network latency between the ACE node and the database nodes.
ACE uses the cluster definition JSON file (opens in a new tab) to connect to nodes and execute SQL statements. It does not use native connection pooling except when initialising a database connection before forking a process. This is because psycopg connections are not process-safe. Therefore, it might be desirable to set up a connection pooler like pgBouncer or pgCat separately and point to that in the cluster JSON file. This may provide faster runtime performance.
ACE cannot be used on a table without a primary key, because primary keys are the basis for range partitioning, hash calculation and several other critical functions in ACE.
If the tables across nodes are vastly different, ACE caps the maximum number of allowed differences to 104 rows. If ACE encounters 10,000 differences, it will terminate early and warn you that it is reaching the maximum differences allowed. This guardrail has been added to prevent ACE from consuming CPU resources when a backup-and-restore might be needed to bring the nodes back in synchrony.
Installing ACE
ACE is installed by the pgEdge Platform installer. The following commands describe installing ACE on a management system (that is not a member of a replication cluster):
-
Navigate to the directory where ACE will be installed.
-
Invoke the pgEdge installer in that location with the command:
python3 -c "$(curl -fsSL https://pgedge-download.s3.amazonaws.com/REPO/install.py)
-
Create a directory named
clusterin thepgedgedirectory created by the pgEdge installer. -
Create and update a .json file, and place the file in
cluster/cluster_name/cluster_name.jsonon the ACE host. For example, if your cluster name isus_eu_backend, the cluster definition file for this should be placed in/pgedge/cluster/us_eu_backend/us_eu_backend.json. The .json file must:- Contain connection information for each node in the cluster in the node's stanza.
- Identify the user that will be invoking ACE commands in the
db_userproperty. This user must also be the table owner.
After ensuring that the .json file describes your cluster connections and identifies the ACE user, you're ready to use ACE commands.
ACE Commands
To review online help about the ACE commands and syntax available for your version, use the command:
./pgedge ace [command_name] --help
Note that the user that invokes ACE commands must be the table owner and specified in the db_user property in the cluster's .json configuration file.
The commands that follow are currently supported by the ACE module.
ACE table-diff
Use the table-diff command to compare the tables in a cluster and produce a csv or json report showing any differences. The syntax is:
$ ./pgedge ace table-diff cluster_name schema.table_name --block_rows=sample_size --max_cpu_ratio=%_cpus --output=json|csv
cluster_nameis the name of the pgEdge cluster in which the table resides.schema.table_nameis the schema-qualified name of the table that you are comparing across cluster nodes.--outputspecifies the output type;choose fromjsonorcsvwhen including the--outputparameter to select the output type for a difference report. By default, the report written todiffs/<YYYY-MM-DD>/diffs_<HHMMSSmmmm>.json. If the output mode is CSV, then ACE will generate coloured diff files to highlight differences.--nodesspecifies a comma-delimited subset of nodes on which the command will be executed. ACE allows up to a three-way node comparison. Simultaneously comparing more than three nodes at once is not recommended.-quiet-mode=Truesuppresses messages about sanity checks and the progress bar instdout. If ACE encounters no differences, ACE will exit without messages. Otherwise, it will print the differences to JSON instdout(without writing to a file).--diff_filespecifies a file that contains mismatched blocks identified during a table comparison. You can specify the file name when re-invoking thetable-diffcommand to recheck a table for mismatches. When you use a--diff_filein conjunction with thetable-diffcommand, the entire block identified in the file is checked; in contrast, thetable-reruncommand targets a specific row within a block, and so the recheck is performed on fewer rows.
The following runtime options can impact ACE performance during a table-diff:
--block_rowsspecifies the number of tuples to be used at a time during table comparisons. ACE computes an MD5 sum on the full chunk of rows per block and compares it with the hash of the same chunk on the other nodes. If the hashes match up between nodes, then ACE moves on to the next block. Otherwise, the rows get pulled in and a set difference is computed. Ifblock_rowsis set to1000, then a thousand tuples are compared per job across tables. It is worth noting here that while it may appear that larger block sizes yield faster results, it may not always be the case. Using a larger block size will result in a speed up, but only up to a threshold. If the block size is too large, the Postgres array_agg() (opens in a new tab) function may run out of memory, or the hash might take longer to compute, thus annulling the benefit of using a larger block size. The sweet spot is a block size that is large enough to yield quicker runtimes, but still small enough to avoid the issues listed above. ACE enforces that block sizes are between 10^3 and 10^5 rows.batch_size: Dictates how many sets ofblock_rowsa single process should handle. By default, this is set to1to achieve the maximum possible parallelism – each process in the multiprocessing pool works on one block at a time. However, in some cases, you may want to limit process creation overheads and use a larger batch size. We recommend you leave this setting to its default value, unless there is a specific use-case that demands changing it.--max_cpu_ratiospecifies the percentage of CPU power you are allotting for use by ACE. A value of1instructs the server to use all available CPUs, while.5means use half of the available CPUs. The default is.6(or 60% of the CPUs). Setting it to its maximum (1.0) will result in faster execution times. This should be modified as needed.
ACE repset-diff
Use the repset-diff command to loop through the tables in a replication set and produce a csv or json report showing any differences. The syntax is:
$ ./pgedge ace repset-diff cluster_name repset_name
cluster_nameis the name of the cluster in which the replication set is a member.repset_nameis the name of the replication set in which the tables being compared reside.--max_cpu_ratiospecifies the percentage of CPU power you are allotting for use by ACE. A value of1instructs the server to use all available CPUs, while.5means use half of the available CPUs. The default is.6(or 60% of the CPUs).--block_rowsspecifies the number of tuples to be used at a time during table comparisons. Ifblock_rowsis set to1000, then a thousand tuples are compared per job across tables.--outputspecifies the output type;choose fromjsonorcsvwhen including the--outputparameter to select the output type for a difference report. By default, the report written todiffs/<YYYY-MM-DD>/diffs_<HHMMSSmmmm>.json.--nodesspecifies a comma-delimited list of nodes on which the command will be executed.
ACE schema-diff
Use the schema-diff command to compare the schemas in a cluster and produce a csv or json report showing any differences. The syntax is:
$ ./pgedge ace schema-diff cluster_name node_one node_two schema_name --output=json|csv
cluster_nameis the name of the cluster in which the table resides.node_oneis the name of the node on which the schema you are comparing resides; you will be comparing the schema to the same schema onnode_two.schema_nameis the name of the schema you will be comparing.outputspecifies the output type;choose fromjsonorcsvwhen including the--outputparameter to select the output type for a difference report. By default, the report written todiffs/<YYYY-MM-DD>/diffs_<HHMMSSmmmm>.json.
ACE spock-diff
Use the spock-diff command to compare the meta-data on two cluster nodes, and produce a csv or json report showing any differences. The syntax is:
$ ./pgedge ace spock-diff cluster_name node_one node_two --output=json|csv
cluster_nameis the name of the cluster in which the table resides.node_oneis the name of the node you will be comparing tonode_two.outputspecifies the output type;choose fromjsonorcsvwhen including the--outputparameter to select the output type for a difference report. By default, the report written todiffs/<YYYY-MM-DD>/diffs_<HHMMSSmmmm>.json.
ACE table-repair
Use the table-repair command to correct any discrepancies or inconsistencies identified by ACE. The syntax is:
$ ./pgedge ace spock-diff cluster_name diff_file source_of_truth schema.table_name --dry-run=false --upsert_only --generate_report
cluster_nameis the name of the cluster in which the table resides.diff_fileis the path and name of the file that contains the table differences.source_of_truthis the name of the node that contains the table that has been manually confirmed to contain accurate rows. The table on other nodes will be updated to match the content of this table.schema.table_nameis the schema-qualified name of the table that you are comparing across cluster nodes.--dry_run- Include this option to perform a test application of the differences between the source_of_truth and the other nodes in the cluster. The default isfalse.--upsert_only(or-u) - Set this option totrueto specify that ACE should make only additions to the non-source of truth nodes, skipping anyDELETEstatements that may be needed to make the data match. This option does not guarantee that nodes will match when the command completes, but can be usful if you want to merge the contents of different nodes. The default value isfalse.--generate_report(or-g) - Set this option to true to generate a .json report of the actions performed;
Reports are written to files identified by a timestamp in the format:reports/<YYYY-MM-DD>/report_<HHMMSSmmmm>.json. The default isfalse.
Example: table-repair Report
{
"time_stamp": "08/07/2024, 13:20:19",
"arguments": {
"cluster_name": "demo",
"diff_file": "diffs/2024-08-07/diffs_131919688.json",
"source_of_truth": "n1",
"table_name": "public.acctg_diff_data",
"dbname": null,
"dry_run": false,
"quiet": false,
"upsert_only": false,
"generate_report": true
},
"database": "lcdb",
"changes": {
"n2": {
"upserted_rows": [],
"deleted_rows": [],
"missing_rows": [
{
"employeeid": 1,
"employeename": "Carol",
"employeemail": "[email protected]"
},
{
"employeeid": 2,
"employeename": "Bob",
"employeemail": "[email protected]"
}
]
}
},
"run_time": 0.1
}Within the report:
time_stampdisplays when the function was called.argumentslists the syntax used when performing the repair.databaseidentifies the database the function connected to.runtimetells you how many seconds the function took to complete.
The changes property details the differences found by ACE on each node of your cluster. The changes are identified by node name (for example, n2) and by type:
- The
upserted_rowssection lists the rows upserted. Note that if you have specifiedUPSERT only, the report will include those rows in themissing_rowssection. - The
deleted_rowssection lists the rows deleted on the node. - The
missing_rowssection lists the rows that were missing from the node. You will need to manually add any missing rows to your node.
ACE table-rerun
If you suspect that the differences reported by ACE are merely temporary (possibly caused due to replication lag) you can use ACE to re-run a previous table-diff run to check if the differences still exist. The syntax is:
$ ./pgedge ace table-rerun <cluster_name> schema.table_name --diff_file=/path/to/diff_file.json
cluster_nameis the name of the cluster in which the table resides.diff_fileis the path and name of the file that contains the table differences.schema.table_nameis the schema-qualified name of the table that you are comparing across cluster nodes.
The table-rerun option reads the diffs from the provided diff file and rechecks the tables to see if those tuples still differ. If ACE identifies any differences, it writes them to a new diff file and reports the file path.